De Novo antibody design using Generative AI and (discrete) flow matching — Patent Published!
Designing functional proteins is a core challenge in computational biology due to the astronomical sequence space. Traditional experimental trial-and-error is now being accelerated by generative AI, which learns from known structures to propose novel biologics that meet specific functional and structural constraints.
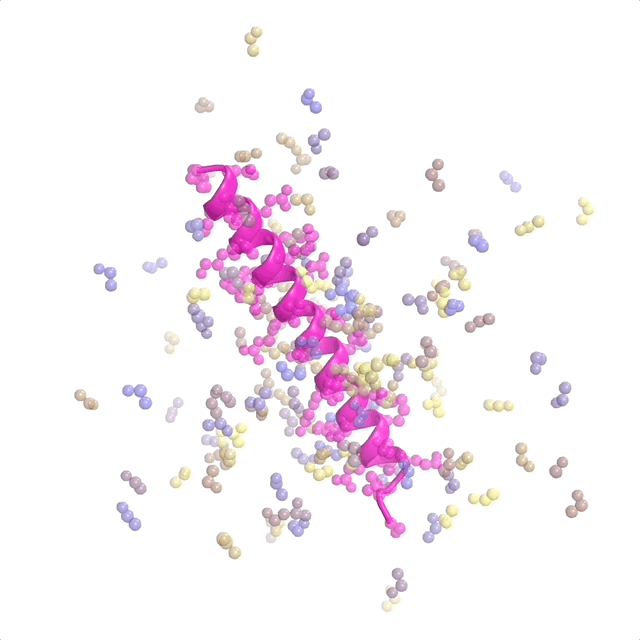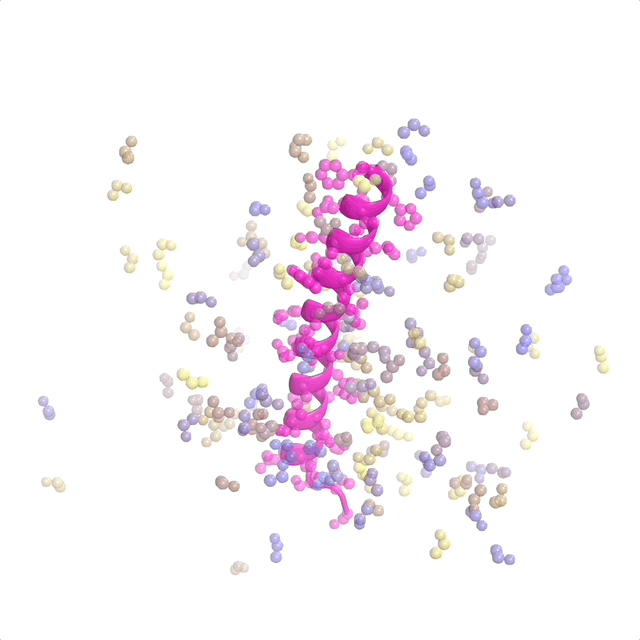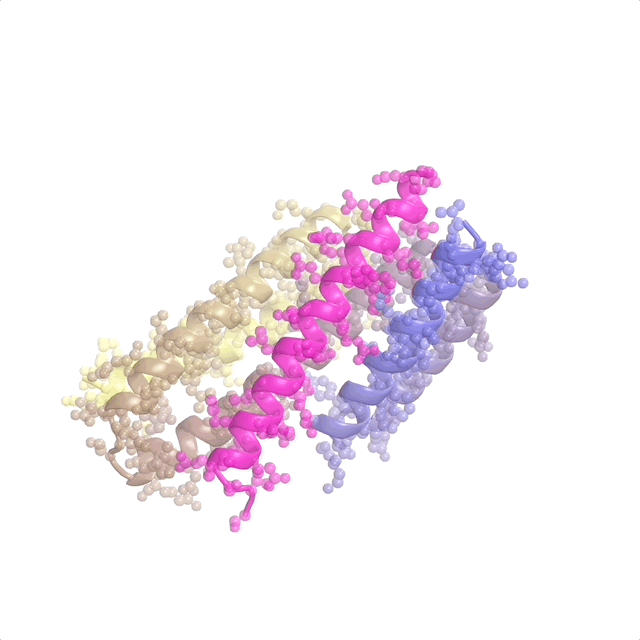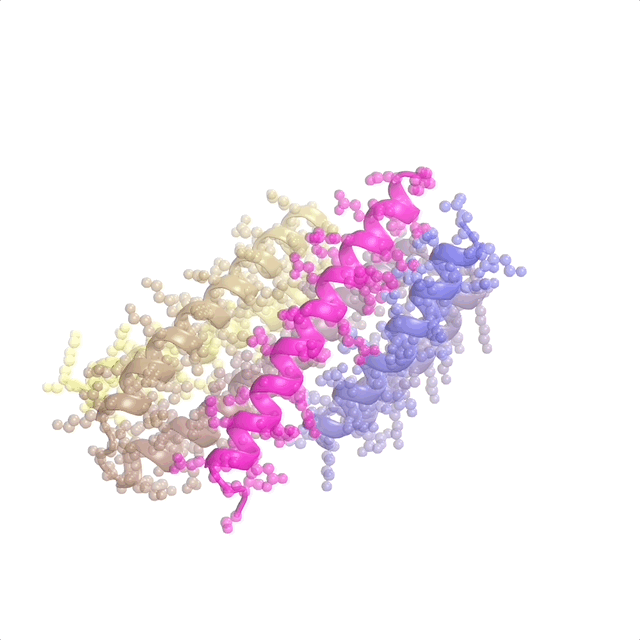
The recently published patent US20240371462A1—Systems and methods for generative design of custom biologics—introduces a framework for generating de-novo peptide backbones and sequences tailored to therapeutic targets, addressing the inverse protein design problem at scale.
This represents pioneer work as the first application of flow matching to protein design. I contributed to discrete flow matching, marking the first use of this technique for the inverse folding problem (designing sequences from binder structures). My work focused on optimizing architectures and training pipelines to ensure predictions are biologically viable and aligned with design goals. This patent underscores how foundational ML translates into impactful tools for biologics discovery and protein science.